Applications of Sensocell optical tweezers
Our technology, allows scientists to probe forces and mechanics in living systems with unprecedented accuracy. Browse below to explore several key applications of optical tweezers in the fields of mechanobiology, molecular biophysics, liquid-liquid phase separation and soft matter physics. We work close with our customers to help them develop their specific applications. If you cannot find your specific field of study in this list, contact us and let’s discuss what we can do!
Cell Mechanobiology
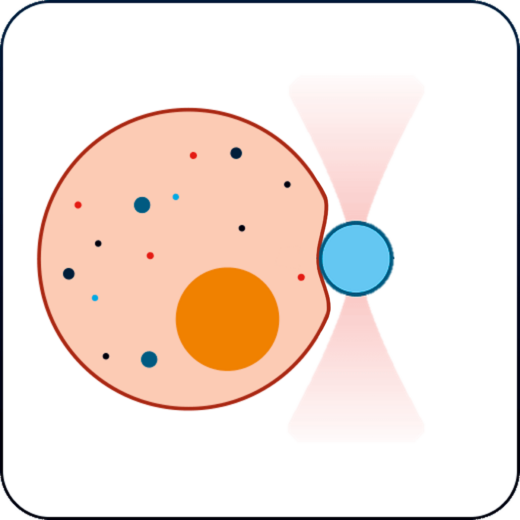
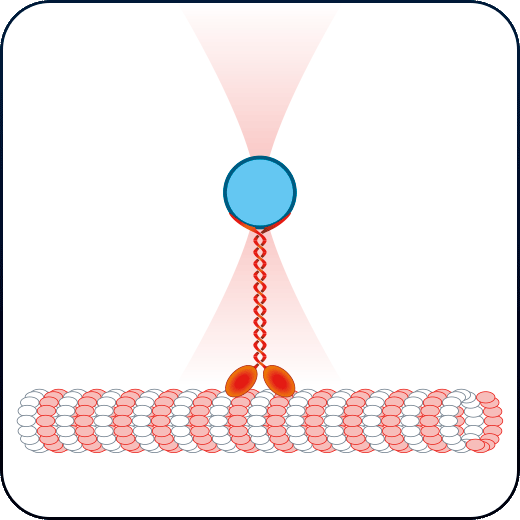
Membrane tension & receptor forces
Study the physical properties of membranes and membrane receptors via tether pulling and particle-cell interaction assays.
Nucleus and subcellular mechanics
Apply creep & stress-relaxation tests inside cells and obtain the mechanical properties of the nucleus and the cytoplasmic medium.
Intracellular rheology
Measure the viscoelastic properties of the cytoplasm and other intracellular medium like the cell nucleus with our automatized active rheology routine.
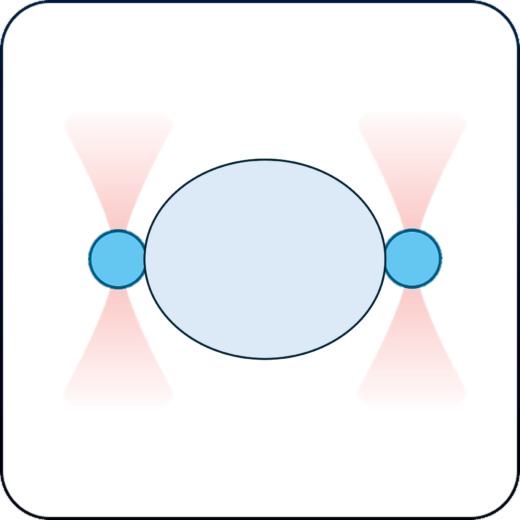
Cell-cell interactions
Establish cell-cell interactions and measure their binding forces while having absolute control on cells orientation and contact time.
Membrane stiffness and elasticity
Study how cells respond to mechanical stress by indenting or stretching cells and characterize their stiffness, elasticity, and viscosity.
Cell motility force dynamics
Measure the forces generated by and exerted on moving cells, and study the dynamics of cell motility.
Membrane tension & receptor forces
Investigate the physical characteristics of membranes and membrane receptors through tether pulling and particle-cell interaction experiments.
Nucleus and subcellular mechanics
Conduct creep and stress-relaxation tests within cells to determine the mechanical properties of the nucleus and cytoplasmic environment.
Intracellular rheology
Assess the viscoelastic properties of the cytoplasm and other intracellular components, such as the nucleus, using our automated active rheology technique.
Cell-cell interactions
Establish and measure the binding forces in cell-cell interactions, with precise control over cell orientation and contact duration.
Membrane stiffness and elasticity
Examine cellular responses to mechanical stress by indenting or stretching cells, and characterize their stiffness, elasticity, and viscosity.
Cell motility force dynamics
Quantify the forces involved in cell movement and analyze the dynamics of cell motility.
Molecular Biophysics
Phase Separation (LLPS)
Colloids & Soft Matter Physics
Motor proteins & filaments
Study the activity of motor proteins and characterize the mechanical properties of microfilaments and microtubules in vitro and in vivo conditions.
Single-molecule interactions
Page not ready… yet.
Please Contact us to know more about this application field.
LLPS of biomolecular condensates
Track changes in the mechanical properties of protein condensates over time by measuring their G modulus with our active rheology routines.
Colloids and soft matter physics
Measure colloidal particles interactions and the rheological properties of colloids, gels, biofilms or liquid crystals.
Molecular Biophysics
Motor proteins & filaments
Investigate motor protein activity and characterize the mechanical properties of microfilaments and microtubules under both in vitro and in vivo conditions.
Single-molecule interactions
Page not ready… yet.
Please Contact us to know more about this application field.
Phase Separation (LLPS)
LLPS of biomolecular condensates
Monitor the evolving mechanical properties of protein condensates by measuring their G modulus using our active rheology techniques.
Colloids & Soft Matter Physics
Colloids and soft matter physics
Assess interactions among colloidal particles and measure the rheological properties of colloids, gels, biofilms, or liquid crystals.
Is your application not listed above? Write us and let’s see what we can do!
Would you like a DEMO?
Download SENSOCELL brochure