Cell membrane tension and cell receptor forces
Study the physical properties of cell membranes by measuring cell membrane tension or cell membrane receptor forces via tether pulling and other particle-cell interaction assays.
Measuring cell membrane tension and receptor forces with optical tweezers
Measuring tension of a cell membrane or quantifying receptor-ligand interactions typically involves the usage of synthetic microspheres. Whether it is a membrane tether pulling assay or a bead-cell adhesion force measurement, the following steps are necessary:
- First, a small bead (usually made of polystyrene or silica) is coated with ligands that can specifically bind to receptors on the cell membrane.
- Then, the bead is optically trapped using the focused laser beam of the optical tweezers.
- Finally, the trapped bead is brought into contact with the cell membrane, where it binds to the membrane receptors via the ligands on its surface. For some cell lines, tethers form naturally and experiments do not need any coating of the beads.
Cell membrane tether pulling assays
Membrane tether pulling experiments allow for local measurement of cell membrane tension. Once the bead attaches to the membrane, optical tweezers pull the bead away from the cell surface, creating a tether that extends the cell membrane outward. The SENSOCELL optical tweezers measure the force applied to the bead and the displacement with high precision. As the bead is pulled, the force required to extend the membrane tether provides information about the membrane’s mechanical properties. Consequently, tension in the cell membrane can be derived from the force-displacement data. Moreover, generating multiple simultaneous tethers on the same cell surface allows the study of force propagation across the membrane.
Cell membrane receptor ligand interaction forces
The SENSOCELL optical tweezers can control very precisely the position of a trapped bead, approaching it to the cell surface undergoing a predefined trajectory or pulling it away from the cell membrane after binding. The system continuously records force and displacement data, enabling the measurement of bond strengths and the dynamics of receptor-ligand interactions.
Selected publications:
- Ravi Das, Li-Chun Lin, Frederic Català-Castro, Nawaphat Malaiwong, Neus Sanfeliu, Montserrat Porta-de-la-Riva, Aleksandra Pidde, Michael Krieg. An asymmetric mechanical code ciphers curvature-dependent proprioceptor activity. Science Advances 17 (2021)
- Andreu, I., Falcones, B., Hurst, S. et al. The force loading rate drives cell mechanosensing through both reinforcement and cytoskeletal softening. Nat Commun 12, 4229 (2021)
- Kechagia, Z., Sáez, P., Gómez-González, M. et al. The laminin–keratin link shields the nucleus from mechanical deformation and signalling. Nat. Mater. 22, 1409–1420 (2023)
Cell membrane tension gradients drives Ca2+ ion channel activity in neurons
This example from the Neurophotonics and Mechanical Systems Biology Lab at the Institute of Photonic Sciences (ICFO) investigates the mechanics of proprioception in Caenorhabditis elegans. The researchers discovered that proprioceptors, sensory mechanoreceptors responding to body movements and positions, activate under compressive stresses in both vivo and in vitro experiments.
Membrane tether pulling with Sensocell optical tweezers
In their in vitro experiments, the authors used SENSOCELL to optically trap microspheres and extrude membrane nanotubes from isolated DVA neuron axons. Using custom pulling routines with the LightAce software SDK, they controlled bead displacement and pulling velocity, applying positive and negative tension gradients to the isolated axons. Simultaneously, they imaged the resultant Ca2+ ion channel activity using confocal microscopy. They observed that Ca2+ signal increments preferentially occurred during the relaxation phase of the tether pulling force-distance curves, indicating that negative tension gradients can induce neuronal activity similarly to axon compression in vivo tests.
Related applications:
Video Confocal video showing a DVA neuron Ca2+ ion channel activity in a dynamic optical trapping assay. The pulling rate applied to the membrane tether is progressively increased. Scale bar = 5µm. Acquired at 10Hz.
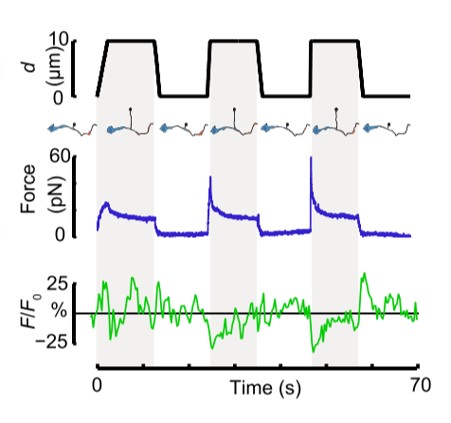
Fig. 1 Time dependence of trap position (top), force signal (middle) and Ca2+ fluorescence signal (bottom) recorded during a membrane tether pulling experiment. The force and Ca2+ ion channel activity signals exhibit negative correlation.
Cell membrane tension propagation revealed via dual tether pulling assays
Multiple tether pulling assays allow studying force propagation across the cell membrane. In this example, two optically trapped beads form two membrane tethers on a neuron axon. Initially, the beads are still and close to the neuron axon. The first tether is then pulled by moving bead 1 several microns away from the axon at high speed. After a few seconds, the force signal relaxes, and bead 1 returns to its original position. The process is repeated for bead 2. Then, bead 2 is moved upwards, several microns away from bead 1, and the previous steps are repeated for both beads.
If the tethers are close enough, data reveals tension propagation across the membrane, seen in the force peak in trap 2 when only the first tether is pulled and vice versa. However, when the tethers are far apart, this phenomenon disappears. Thus, tuning the tether-to-tether distance quantifies how cell membrane tension decays with distance.
Fig.1 Confocal video of the described dual tether pulling assay performed on a neuron axon. Courtesy of M. Krieg lab (ICFO).
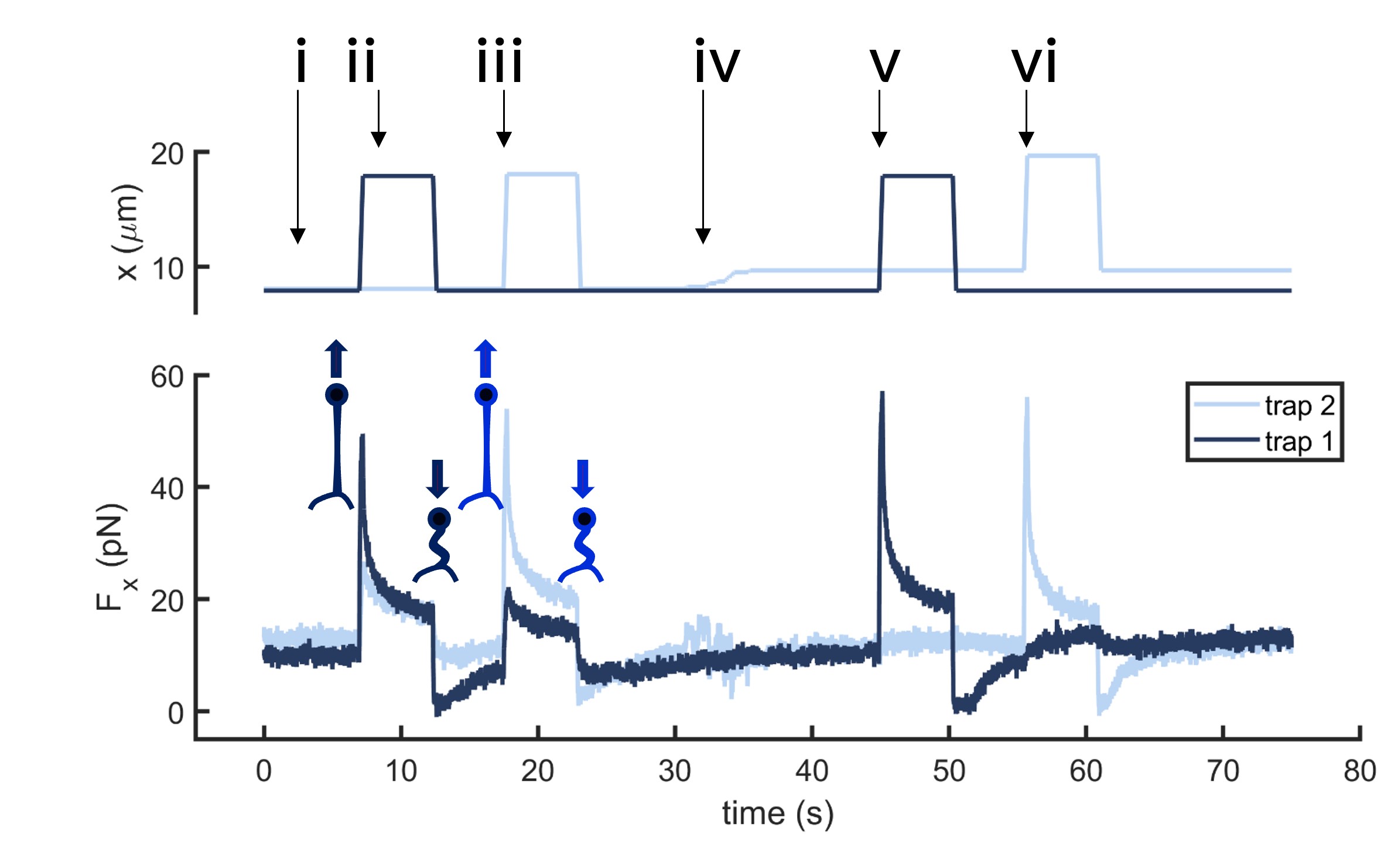
Fig.2 Time dependence of trap 1 and trap 2 position (top) and force (bottom) data along the different steps of the described dual tether pulling experiment.
Force loading rate on the cell membrane drives the maturation of single adhesions
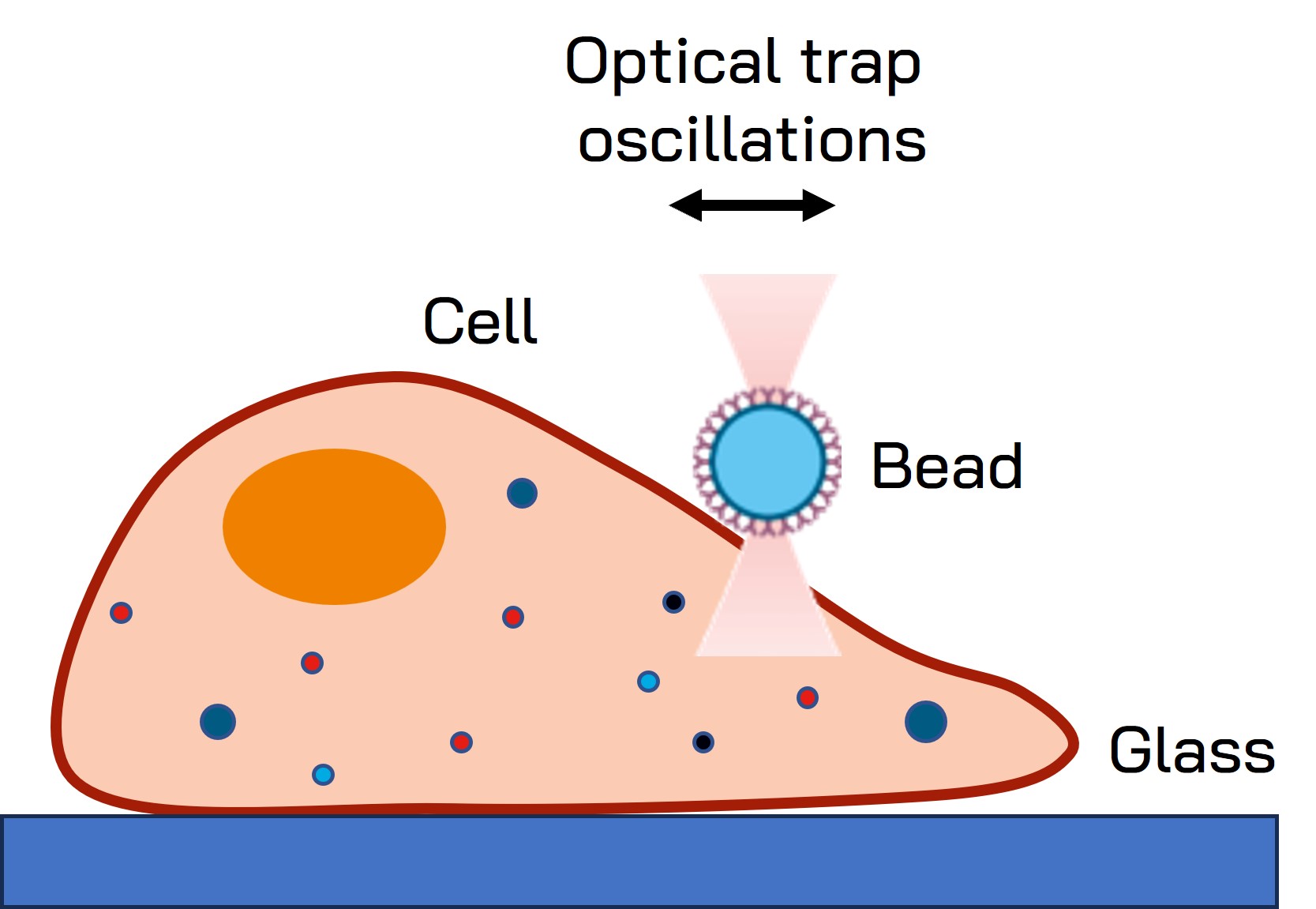
Fig. 1 Schematic representation of the optical tweezers experiment. A fibronectin-coated bead is trapped, brought into contact with the cell membrane and stimulated with oscillations at different frequencies.
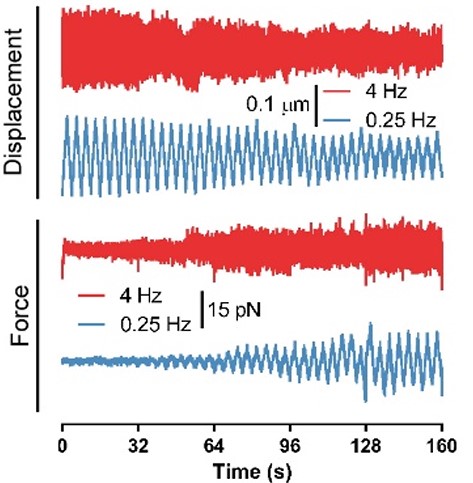
Fig. 2 Force and displacement data for two beads stimulated at frequencies of 4 Hz and 0.25 Hz. The progressive reduction of the beads displacement is in concomitancy with an increase of the applied force and loading rate.
Key Concepts
- Tether pulling experiments: These involve attaching a bead to the cell membrane and using optical tweezers to pull a tether (a thin membrane tube). This allows the study of membrane mechanics, including elasticity and viscosity.
- Membrane tension: typically, the initial slope of the force versus displacement curve provides the membrane tension. The relationship between the force (F), the tether radius (R), and the membrane tension (σ) can be described by the formula F=2πRσF
- Membrane receptor force measurements: optical tweezers can apply controlled forces to membrane receptors, enabling the study of how these receptors respond to mechanical stress and how force influences receptor-ligand interactions.
Advantages
- Precision: optical tweezers provide extremely precise control over the position and force applied to particles, allowing detailed measurement of cellular and molecular responses.
- Quantitative data: the ability to measure forces in the picoNewton range and displacements in the nanometer range enables quantitative analysis of membrane mechanics and receptor behavior.
- Non-destructive: optical tweezers can manipulate cells and molecules without physical contact, minimizing damage and preserving cell viability.
- Dynamic measurements: the variety of trap motion predefined and customizable routines integrated in the SENSOCELL system allow real-time monitoring of mechanical properties and responses to applied forces, providing insights into dynamic cellular processes.
- High spatial resolution: optical tweezers assays can target specific regions of the cell membrane or individual receptors, enabling localized measurements and manipulation.
Conclusions
The SENSOCELL optical tweezers offer a powerful and precise method for studying the mechanical properties of cell membranes and the force-dependent behavior of membrane receptors.
Would you like a DEMO?
Download SENSOCELL brochure
